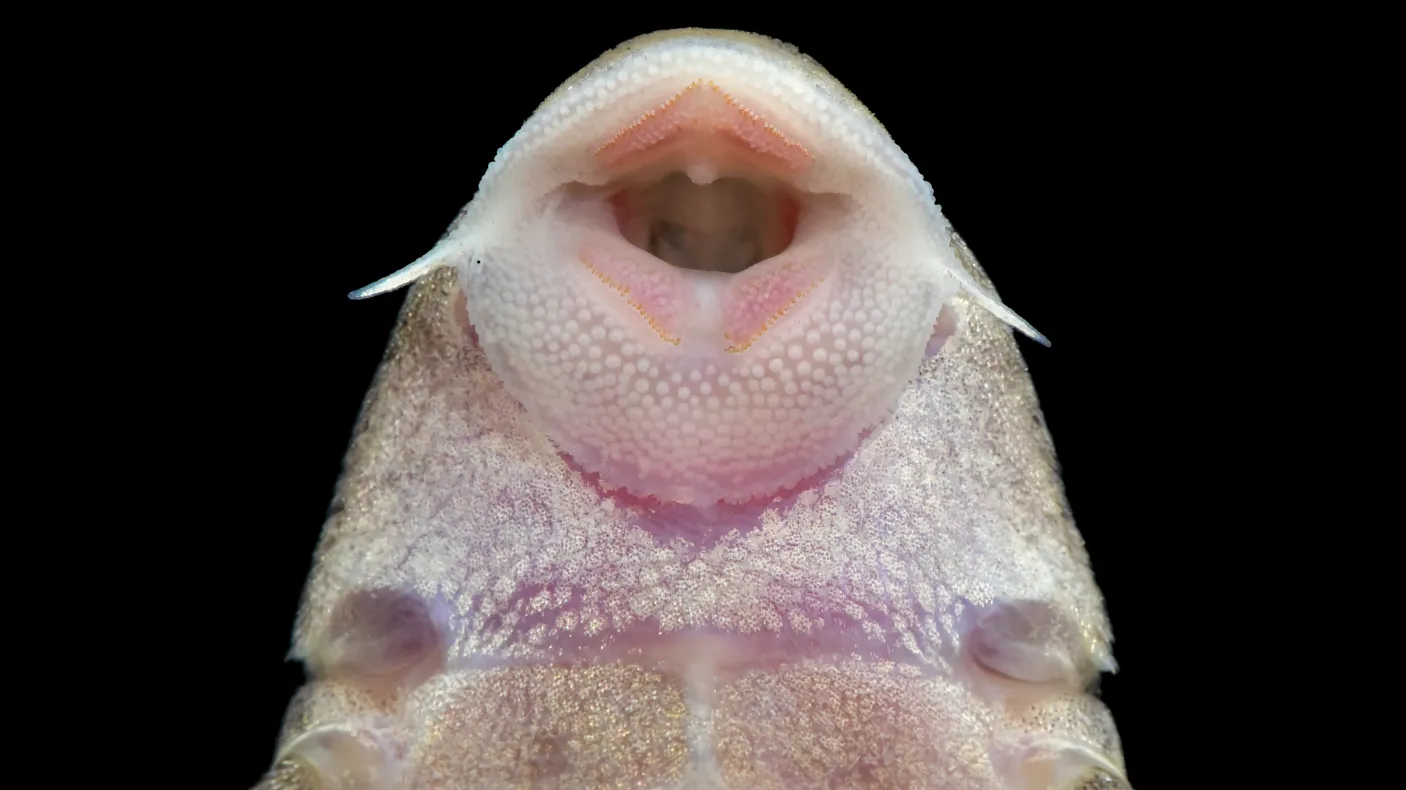
What’s in a Name?
Published
Categories
Author
How many of us, upon seeing a bird
How many of us, upon seeing a bird, plant, or insect, stop to think … Who is that? How should it be classified? What species is that? Fundamental to humanity’s relationship with nature is our desire to classify and name our non-human neighbours, with local names varying widely by region and culture. Indeed, how we answer these questions says a lot about where, and with whom, we live.
The common lake fish known in Ontario as Sheepshead goes by Gaspergoo in Louisiana. In Toronto, any small silvery fish might be called a “minnow,” whereas aficionados and scientists differentiate among no fewer than 38 minnow species in Ontario alone, from the Allegheny Pearl Dace to the Spottail Shiner. Taking this even further, Tukano cultures in the northwestern Amazon are noted for their precision in distinguishing each of several hundred tropical fish species inhabiting the streams and rivers of their rainforest home, echoing the famous precision with which Inuit cultures in Canada recognize gradations of snow and ice.
The myriad ways in which people name nature is, on one hand, a poignant manifestation of culturally diverse perspectives. Yet for biodiversity scientists who must accurately and efficiently collect, collate, and communicate species data across generations, cultures, and communities, a parallel system for consistently and precisely naming nature is critical, as are new tools for rapidly obtaining the correct name for each species.
Tragically, the ongoing acceleration of global biodiversity loss is placing scientists and researchers in an urgent race to not only protect documented species but also discover, differentiate, and name the staggering diversity of species that remain undescribed. More so now than ever, biologists need the help of new technologies and big data to rapidly and unambiguously assign Earth’s multitude of species their proper scientific name. ROM’s Natural History curators and collections, and the cutting-edge biodiversity research they lead and facilitate, are helping pave the way forward.
Much of my recent research as ROM’s Associate Curator of Fishes has focused on resolving taxonomic confusion and improving species identification tools for fishes in the Western Amazon of eastern Ecuador and northern Peru. Rivers in this area are the most biodiverse in the Amazon basin, which itself is Earth’s most biodiverse freshwater ecosystem. If fisheries biologists in Ontario find it challenging to identify a few dozen minnow species—a fraction of the estimated 5,000 fish species in the Amazon—revolutionary new technologies will be required for local and foreign scientists and conservationists to rapidly, yet accurately, identify and monitor Amazonian fish communities facing threats from dams, mining, deforestation, and drought.
Working with eDNA is like working with magic. It is as if we have been teleported to a future Star Trek universe in which any object or organism can instantly be identified with the mere swipe of a tricorder.
Environmental DNA
Environmental DNA (eDNA) metabarcoding* is a state-of-the-art approach that students and staff of ROM’s Fish Division, collaborators in Ecuador and Peru, and I are developing to meet this need. Foundations for this approach were laid only a couple decades ago in Guelph, Ontario, by Professor Paul Hebert, the pioneering founder of the Barcode of Life initiative. Dr. Hebert was inspired while scanning groceries to develop a DNA-based tool for identifying any species on Earth, just as the unique sets of vertical black lines help differentiate whole milk from 2 percent at the grocery store checkout counter. For hundreds of years before then and still today when we’re unable to immediately read a species’ DNA, taxonomic identifications can be made only by examining anatomical characteristics that can be highly variable and require specialized training to properly interpret.
The Barcode of Life initiative launched a global taxonomic renaissance. Labs around the world rushed to sequence the cytochrome oxidase I (COI) gene—the DNA region that Hebert and colleagues designated as a global standard—to build reference databases for as many species as possible. In a remarkably short period, vast libraries of COI sequences for thousands of species were generated, aggregated, and archived in publicly accessible, government-funded repositories.
Unfortunately, cracks in this system have also begun to show. In the decentralized rush to generate and share COI sequences for all Earth’s species, critical links between these sequences and source specimens were lost. Even when such links were retained, the invaluable source specimens that link the COI sequences to our system of taxonomy via their anatomical characters have too often been kept in small, remote, or poorly resourced labs unwilling or unable to make specimens available for ID verification by taxonomic experts, who themselves have too few resources to travel to complete such verifications. Consequently, existing barcode reference libraries are plagued by misidentifications, and correcting these misidentifications on a large scale is nearly impossible.
Now, the advent of eDNA metabarcoding has instigated a second revolution in molecular taxonomy, with astounding implications, especially for detecting rare, endangered, or potentially threatening organisms, such as invasive species. Using new genetic sequencing technologies that enable the eDNA approach, biodiversity scientists have suddenly been empowered to precisely identify hundreds of species at a time from simple environmental samples, such as a few litres of water or even air, based on DNA contained in the multitude of dead cells that all multicellular organisms constantly shed into their environments.
Working with eDNA is like working with magic. It is as if we have been teleported to a future Star Trek universe in which any object or organism can instantly be identified with the mere swipe of a tricorder. While we are still a long way from being able to conduct eDNA analyses instantly, our ability to molecularly identify up to several hundred species in a water sample in a few days is still a major advancement over what previously would have required weeks to months of arduous fieldwork.
Serendipitously, the new metabarcoding approach is also providing biodiversity scientists the much-needed opportunity to build all new specimen-backed barcode reference databases in a way that resolves longstanding flaws in the previous piecemeal approach. Due to limitations of the sequencing technologies used for eDNA, the COI sequence that was Hebert’s original standard has been replaced by a neighbouring gene known as 12S. As a result, entirely new reference databases must be generated for the new barcode region, providing a critical opening that ROM, with its vast collection of frozen genetic samples and linked whole specimens, is ideally positioned to fill.
Researchers in the field
Recognizing this need
Recognizing this need and opportunity in 2021, I was fortunate to be invited by colleagues at the Universidad de las Américas (UDLA) in Quito, Ecuador, to lead the creation of a new eDNA reference database for the Ecuadorian Amazon. I joined collaborators from UDLA and World Wildlife Fund on two expeditions to eastern Ecuador in 2021 and 2022 and led two additional expeditions in 2022 and 2023 to adjacent areas of northern Peru with colleagues from the Universidad Nacional Mayor de San Marcos in Lima and the Instituto de Investigaciones de la Amazonía Peruana in Iquitos, and funding from ROM and the New Orleans-based Coypu Foundation. In total, this fieldwork yielded a collection of nearly 10,000 whole specimens of over 600 Western Amazonian fish species, plus approximately 4,000 frozen genetic samples. My lab in ROM’s Fish Division, with the help of Museum technicians and students from the University of Toronto, is using this material to establish the first centralized, taxonomically comprehensive, 12S DNA reference library for Western Amazonian fishes.
Prior to our project, there was not a single COI sequence for a fish from the Ecuadorian Amazon in a major publicly accessible repository and only one 12S sequence. Already, our project has generated over 400 COI sequences and over eight hundred 12S sequences, and the library is growing.
Our goal is to make a combined 12S and COI reference library that is publicly available and to link each of the species-specific DNA sequences to a specimen that is archived and permanently accessible at ROM. This will mean that when the library is used to identify eDNA samples, future generations of scientists will know exactly to which individual specimen the DNA sequence belongs. Moreover, researchers will be able to either visit ROM or request each specimen on loan to study it and update taxonomic identifications as our understanding of Amazonian diversity and species nomenclature changes over time.
In a recent trial using an early draft of our new reference library to identify real environmental DNA sequences from the Ecuadorian Amazon, our data performed well, improving the rate of accurate species-level identifications by over 60 percent compared with existing non-Ecuadorian databases. The total number of species identified remains low, though, increasing from only 87 to 141 out of a total of approximately 410 fish species indicated by the eDNA samples.
While we are making enormous advancements in novel methodologies to survey biodiversity, building the infrastructure to maximize these tools is an uphill climb. The years of field- and museum-based work required to collect and name each species to establish a comprehensive library of linked specimens and sequence data is a monumental task. Still, this work is essential. At a time when hope for saving nature can be scarce, I am more excited than ever that the tremendous potential of ROM’s century-old Natural History collections can be unlocked to meet the urgent needs presented by modern approaches to naming and protecting our non-human neighbours.
*“Meta” in this context refers to the synchronous reading of hundreds of DNA sequences from a single environmental sample, with a computer needed to disentangle the individual sequences.
Nathan K. Lujan
Nathan K. Lujan is Associate Curator of Fishes at ROM. This position is generously supported by the Herbert A. Fritch Family Foundation.